Congratulations to
Protein map
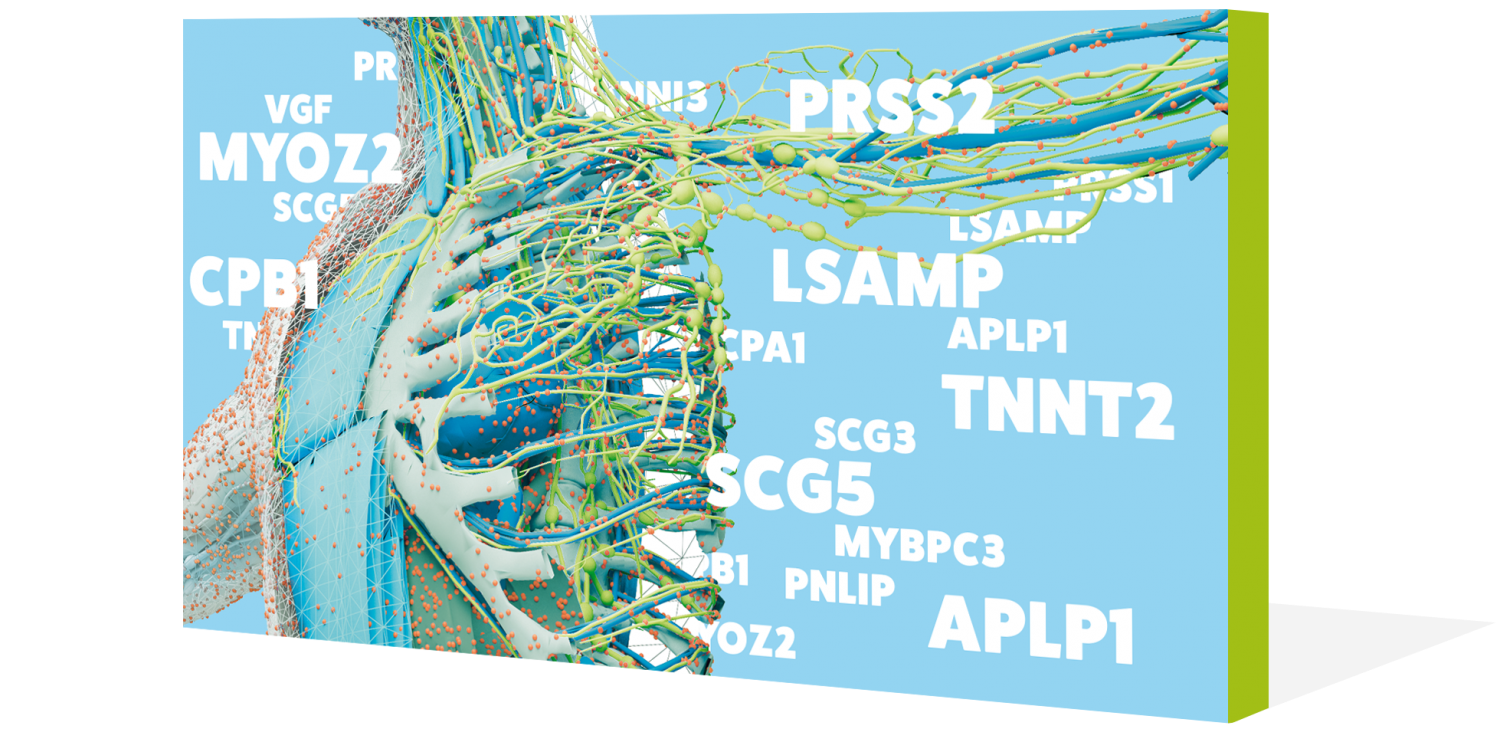
Picture: ediundsepp Gestaltungsgesellschaft, Munich

Picture: ediundsepp Gestaltungsgesellschaft, Munich
They determine what type of hair we have, digest our food and fight against pathogens – nothing would work without proteins in the human body. A research team at the Technical University of Munich (TUM) has identified 18,097 of these molecules and thus produced an almost complete inventory of the human proteome. The aim is to enable individualized treatment for all patients in the future – even for diseases such as cancer.
Scrutinizing saliva, blood and earwax was all part of the quest to identify the multi-talented all-rounders of the human body: proteins. The mission also involved examining a wide variety of body tissues and fluids, and even numerous tumor cells. As a result of all this, in 2014, researchers led by biochemist Bernhard Küster were able to produce the first map of the human proteome – that is, the complete set of proteins inside our bodies. They successfully identified 18.097 of these proteins, which accounts for over 90 percent of the proteome. They also located these proteins within the body and worked out how many variants each one has.
This is an important step in developing personalized drug therapies, tailored specifically to each individual patient. In recent years it has become increasingly clear that not only our genes, but also our proteins have an influence on us and our diseases. This holds particularly true for cancer. Tumors develop from aberrant body cells, which vary greatly from one patient to another. Personalized medicine thus takes account of both the patient’s genetic makeup and their proteome. In the case of cancer, initial investigations by Küster and his research team suggest that the effect of drugs on cancer cells varies between people – precisely due to differences in proteins.
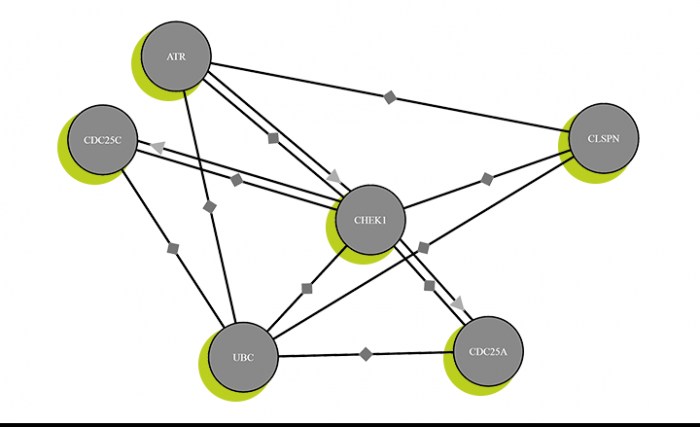
Küster and his team have used their results to create the most comprehensive protein database to date: ProteomicsDB. Newly developed software means they can store, link and evaluate their data – as can other protein experts all over the world. Anyone can add the results of their own protein research, since the program is freely available on the Internet. In this way, all of the contributors are gradually filling in the last blanks on the human protein map.
Proteins are real masters of transformation, so analyzing them is extremely difficult. Depending on where they occur in the body and in what conditions, they slip into different roles, changing their chemical properties and appearance – which makes them even harder to differentiate. So for each of the almost 20,000 proteins identified by Küster and his team, there are still countless variants. The task facing researchers in the future is to find them all.

Since proteins are tiny, advanced analytical techniques are required to study them. One such technique is mass spectrometry. Here, a protein’s molecular chain is first broken down into short fragments, which are sorted according to weight and electrical charge. The data obtained from these individual pieces can then be evaluated and virtually reassembled by computer – similar to doing a jigsaw puzzle. By comparing this data with established data sets, researchers can then determine very precisely which protein is involved. This process thus depends on large databases containing all known proteins – such as the ProteomicsDB set up by Bernhard Küster.
“If we can establish the protein profile of a tumor, for instance, we could administer drugs in a much more selective way in the future.”

Bernhard Küster, Professor of Proteomics and Bioanalytics at TUM
Picture: Filser